Depression Preprint Analysis, Part 1
This is the first post in a series focused on trying to analyze the contents of a collection of preprint papers on a topic – in this case, depression. This post involves how I scraped the (initial) website, along with some analysis of basic information from the descriptions of the preprints.
OSF
The OSF (Open Science Foundation) is project by the Center for Open Science, dedicated to aggregating the results of multiple arXiv-like preprint services and making them searchable as a group. As a result, it makes for a potentially interesting jumping off point to get preprints from a number of different sources.
The interface for the search is pretty standard, and the site returns abstracts and other basic information on the preprints themselves, along with links to the preprints on their respective sites.
The data in these summaries is pretty well-structured, so getting the information out of the respective elements with BeautifulSoup isn’t too bad. That said, the returned results are rendered with JavaScript, so you need to render the page fully before scraping it. As a result, we also need Selenium to fully automate the process.
Scraping the OSF Website
First, library imports and initial setup:
>> from collections import namedtuple
>> from time import sleep
>>
>> from bs4 import BeautifulSoup
>> import pandas as pd
>> from selenium import webdriver
>> from tqdm import tqdm ## just for monitoring purposes, not necessary for functionality
>>
>> ## initial URL for search results
>> SEARCH_URL = "https://osf.io/preprints/discover?q=depression"
>>
>> ## for storing results
>> OSFPaperTuple = namedtuple("OSFPaperTuple",
>> field_names=["PaperName","URL","LastEdited","Categories","Source"])
>>
>> driver = webdriver.Firefox()
>> driver.get(SEARCH_URL)Identifying the pages is fairly simple, all pages of the search can be accessed via a URL of the form https://osf.io/preprints/discover?page=XXX&q=depression. Getting the number of pages returned in the search is necessary, but this is fortunately included in an element at the bottom of the page.
>> page_html = BeautifulSoup(driver.page_source, "lxml")
>> pagination_element = page_html.find_all("ul", attrs={"class":"pagination"})[-1]
>> last_page_link = pagination_element.find_all("li", attrs={"class":"ember-view"})[-1]
>> number_of_pages = int(last_page_link.text.strip())We also need to extract the information of interest from each result: the title of the preprint, the URL to its location, the date of the last edit, the categories that the preprint is labeled with, and the name of the source which is hosting the preprint. The search results are almost totally consistent – the only exception is that some preprints are missing URL links, but those can be detected by checking for the presence of an <a> element in the title element.
>> def get_OSF_paper_info(paper_element):
>> ## Collects information on the papers and returns them in a named tuple.
>> paper_link = paper_element.find("h4")
>> if paper_link.a:
>> paper_name = paper_link.a.text.strip()
>> paper_url = paper_link.a["href"]
>> else:
>> paper_name = paper_link.span.text.strip()
>> paper_url = ""
>>
>> last_edited = paper_element.find("em").text.strip()[13:-4]
>>
>> category_elements = paper_element.find_all("span", attrs={"class":"subject-preview"})
>> if category_elements:
>> categories = [e.text.strip() for e in category_elements]
>> else:
>> categories = []
>>
>> source_name = paper_element.find("span", attrs={"class":"search-result-providers"}).text.strip()
>> return OSFPaperTuple(paper_name, paper_url, last_edited, categories, source_name)Putting this all together is fairly simple:
>> paper_tuples = []
>> for page in tqdm(range(number_of_pages)):
>> target_page = f"https://osf.io/preprints/discover?page={page+1}&q=depression"
>> driver.get(target_page)
>> sleep(5) ## try to avoid overwhelming the site
>> page_html = BeautifulSoup(driver.page_source, "lxml")
>> papers_on_page = page_html.find_all("div", attrs={"class": "col-sm-8"})[1]
>> paper_elements = papers_on_page.find_all("div", attrs={"class":"ember-view"}, recursive=False)
>> current_page_tuples = [get_OSF_paper_info(p) for p in paper_elements]
>> paper_tuples.extend(current_page_tuples)At the time of running this, there were 443 search pages to scrape. I added a delay as I didn’t want to hit the OSF site too heavily with search page requests, so it took about 40 minutes to complete this.
From there, we can convert the list of named tuples to a dataframe, save those results, and then shut down the Selenium browser.
>> paper_df = pd.DataFrame(paper_tuples)
>> paper_df.to_csv("OSF_paper_info.csv", index=False)
>>
>> driver.close()Analysis
The returned data consists of 4,414 preprints and their accompanying information. Unfortunately, almost a thousand are duplicates, though there are still plenty to look at:
>> # note: I read this in later, so the Categories column is loaded
>> # as a string instead of a list; that's fixed later
>> paper_df = pd.read_csv("OSF_paper_info.csv")
>> len(paper_df)
4414
# there are duplicates, unfortunately
>> paper_df = paper_df[~paper_df.duplicated()]
>> len(paper_df)
3475With this initial data, I’m trying to answer four different questions:
- What are the counts of each source?
- What are the most common word stems in the titles?
- How often do various categories appear?
- How are the “last updated” years distributed?
Question 1: Source Counts
This is easily done:
>> paper_df["Source"].value_counts()
RePEc 1625
PsyArXiv 696
arXiv 433
bioRxiv 297
Preprints.org 159
OSF Preprints 143
SocArXiv 38
PeerJ 28
MindRxiv 15
INA-Rxiv 9
Thesis Commons 9
SportRxiv 7
EarthArXiv 6
MetaArXiv 4
Cogprints 2
AfricArXiv 2
AgriXiv 1
NutriXiv 1
Name: Source, dtype: int64I’ll set aside the sources with fewer than 100 preprints just due to size. Among the remainder:
- RePEc is short for Research Papers in Economics, so it’s focused more on economic depressions that the psychological condition.
- PsyArXiv is focused on “the psychological sciences” as its front page puts it, so it’s likely to be the primary source here.
- arXiv, judging from some of the titles, is using the word “depression” in several contexts, none of which are likely aligned with this analysis:
>> import random >> random.sample(paper_df[paper_df["Source"]=="arXiv"]["PaperName"].tolist(), 6) ['Attractor Dynamics with Synaptic Depression', 'A geometrical height scale for sunspot penumbrae', 'Understanding Forbush decrease drivers based on shock-only and CME-only models using global signature of February 14, 1978 event', 'Spectral shape of the UV ionizing background and HeII absorption at redshifts 1.8 < z < 2.9', 'The nature of the light variability of the silicon star HR 7224', 'Drifting Asteroid Fragments Around WD 1145+017'] - bioRxiv is focused on genetics and biological science, and it might have some relevant preprints:
>> random.sample(paper_df[paper_df["Source"]=="bioRxiv"]["PaperName"].tolist(), 6) ['Inhibition of protein translation by the DISC1-Boymaw fusion gene from a Scottish family with major psychiatric disorders', 'Insight into the genetic architecture of back pain and its risk factors from a study of 509,000 individuals', 'Genetic correlations between pain phenotypes and depression and neuroticism', 'A spike timing-dependent plasticity rule for single, clustered and distributed dendritic spines', 'Glucocorticoid receptor-mediated amygdalar metaplasticity underlies adaptive modulation of fear memory by stress', 'Pharmacogenetics of antidepressant response: a polygenic approach'] - Preprints.org and OSF preprints have somewhat generic names compared to the other sources, and while some of the preprint titles look relevant, they seem like a more esoteric mix:
>> random.sample(paper_df[paper_df["Source"]=="Preprints.org"]["PaperName"].tolist(), 6) ['Development of a Novel Staging Model for Affective Disorders Using Partial Least Squares Bootstrapping: Effects of Lipid-Associated Antioxidant Defenses and Neuro-Oxidative Stress', 'Understanding the Demographic Predictors and Association of Comorbidities in Hospitalized Children with Conduct Disorder', 'Effect of a Comprehensive Health Care Program on Blood Pressure, Blood Glucose, Body Composition, and Depression in Older Adults Living Alone: A Quasi-experimental Pre-posttest Study', 'Impact of Service User Video Presentations on Explicit and Implicit Stigma toward Mental Illness among Medical Students in Nepal: A Randomized Controlled Trial', 'Interpolation of Small Datasets in the Sandstone Hydrocarbon Reservoirs, Case Study from the Sava Depression, Croatia', 'Transpersonal Gratitude, Emotional Intelligence, Life Contentment, and Mental Health Risk Among Adolescents and Young Adults'] >> random.sample(paper_df[paper_df["Source"]=="OSF Preprints"]["PaperName"].tolist(), 6) ['Music and mood regulation during the early-stages of the COVID-19 pandemic', 'High frequency stimulation-induced plasticity in the prelimbic cortex of rats emerges during adolescent development and is associated with an increase in dopamine receptor function', 'Preprint BuckfieldSinclairGlautier (2019) Slow associative learning in alcohol dependence and the Alcohol Cue Exposure Treatment Paradox', 'Subclinical anxiety and depression are associated with deficits in attentional target facilitation, not distractor inhibition', 'Placebo response and psychosis: a putative shared mechanism', 'Combat stress in a small-scale society suggest divergent evolutionary roots for posttraumatic stress disorder symptoms']
As a side note, something largely missing from these titles is the matter antidepressant drugs and other pharmacological topics. I would guess that it’s the result of some level of sampling bias, rather than an accurate cross-section of depression research as a whole.
Question 2: Most Common Word Stems
Investigating common elements of the titles could be illustrative as well. Examining the stems of the words is probably more useful than checking the words themselves – for example, separate counts of “depression” versus “depressive” versus “depressed” wouldn’t add much useful information than just the “depress” root.
>> from collections import Counter
>> import re
>>
>> import nltk
>> from nltk.stem.snowball import SnowballStemmer
>>
>> def title_list_to_stems(title_list, stemmer, stopwords):
>> # note: gotta use .lower(), otherwise stopwords aren't properly caught
>> word_list = [word for title in title_list for word in nltk.word_tokenize(title.lower())]
>> word_list = [word for word in word_list if re.search("[A-Za-z]", word)]
>> word_list = [word for word in word_list if word not in stopwords]
>> stems = [stemmer.stem(word) for word in word_list]
>> return stemsThe above function is just used to turn a list of preprint titles into a list of word stems. The code below does this for each of the six preprint sources mentioned above, and determines the 15 most common stems:
>> stemmer = SnowballStemmer("english")
>> english_stopwords = set(nltk.corpus.stopwords.words("english"))
>>
>> num_most_common = 15
>> sources_with_enough_papers = ["RePEc", "PsyArXiv", "arXiv", "bioRxiv", "Preprints.org", "OSF Preprints"]
>> paper_stems_dict = {}
>> for s in sources_with_enough_papers:
>> titles = paper_df[paper_df["Source"] == s]["PaperName"].tolist()
>> stems = title_list_to_stems(titles, stemmer, english_stopwords)
>> most_common_stems = Counter(stems).most_common(num_most_common)
>> paper_stems_dict[s] = [m[0] for m in most_common_stems]
>>
>> pd.DataFrame(paper_stems_dict)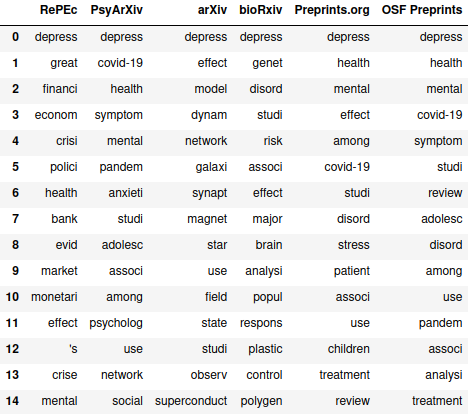
Unsurprisingly, “depress” is at the top for all six sources. As you could probably guess from the sampling of the preprint titles earlier, RePEc’s stems are more economics-focused, arXiv is kind of a mixed bag (and isn’t really anything we’re looking for), and the other four are focused on the psychological condition, with bioRxiv focused more on the biology and genetics perspective. Interestingly, COVID-19 actually ends up as the second most common stem in PsyArXiv’s articles and well inside the top 15 for Preprints.org and OSF Preprints. Checking how many times they appear:
>> covid19_mentions = paper_df[paper_df["PaperName"].apply(lambda x: "covid" in x.lower())]
>> covid19_mentions["Source"].value_counts()
PsyArXiv 149
Preprints.org 21
OSF Preprints 19
SocArXiv 11
AfricArXiv 2
arXiv 1
Name: Source, dtype: int64The term “covid” appears in 149 PsyArXiv preprints, which is a little over one-fifth of their preprints in the search. For Preprints.org and OSF preprints, it’s just over one-eighth. It’s interesting to me that the second most common stem for PsyArXiv preprints only appears around 21% of the time – I’m not sure if that’s exceptional or not.
Question 3: Frequencies of Categories
To start this off, I know from looking around at the OSF’s search portal that there were a number of preprints which did not have any categories at all. It actually turns out that the majority of them lack categories:
>> # do this step only if you loaded the file from the CSV - it's read back in as
>> # a string by pd.read_csv()
>> paper_df["Categories"] = paper_df["Categories"].apply(eval)
>> (paper_df["Categories"].apply(lambda x: x == [])).mean()
0.6529496402877698The primary offender here seeps to be RePEc:
>> pd.crosstab(paper_df["Source"], paper_df["Categories"].apply(lambda x: x == []))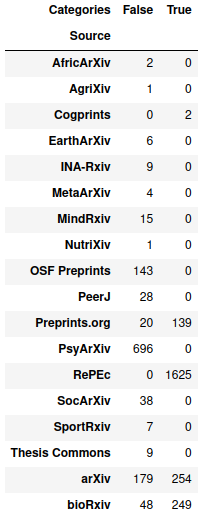
On the plus side, since PsyArXiv and OSF preprints have categories for all their preprints, economics topics don’t end up in the most common categories (though it looks like one topic from arXiv sneaks into 15th place):
>> Counter([x for y in paper_df["Categories"].tolist() for x in y]).most_common(20)
[('Social and Behavioral Sciences', 760),
('Psychology', 673),
('Clinical Psychology', 433),
('Medicine and Health Sciences', 376),
('Medical Specialties', 260),
('Psychiatry', 255),
('Life Sciences', 162),
('Health Psychology', 127),
('Biology', 126),
('Neuroscience and Neurobiology', 120),
('Psychiatry and Psychology', 103),
('Child Psychology', 85),
('Personality and Social Contexts', 78),
('Cognitive Psychology', 77),
('Physics', 75),
('Quantitative Psychology', 66),
('Mental Disorders', 59),
('Social Psychology', 56),
('Developmental Psychology', 45),
('Counseling Psychology', 38)]Question 4: How are the “last updated” years distributed?
Finally, the question of how the preprints are distributed in time. A basic check of the year reveal that there hasn’t exactly been a steady increase:
>> paper_df["LastEdited"] = pd.to_datetime(paper_df["LastEdited"])
>> paper_df["Year"] = paper_df["LastEdited"].dt.year
>> paper_df["Year"].value_counts().sort_index()
2007 40
2008 16
2009 82
2010 11
2011 22
2012 31
2013 176
2014 458
2015 488
2016 638
2017 258
2018 309
2019 138
2020 441
2021 367
Name: Year, dtype: int64There’s a peak in 2016, which no year before or since (or so far, in 2021’s case) has matched. I’m not really sure when some of these preprint services took off, so it’s hard to tell exactly what’s driving this. Looking at a cross-tabulation of the six sources we looked at before versus year:
>> papers_with_good_sources = paper_df[paper_df["Source"].isin(sources_with_enough_papers)]
>> pd.crosstab(papers_with_good_sources["Year"], papers_with_good_sources["Source"])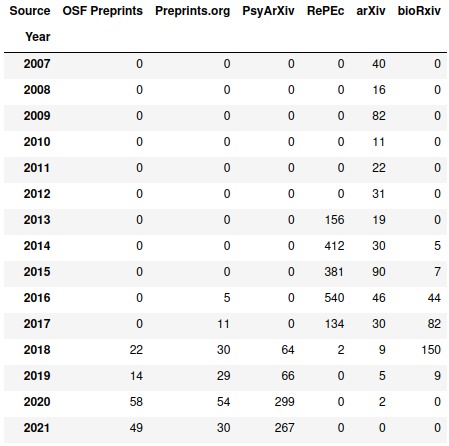
So basically all of the spike appears to come from RePEc. It’s interesting that it seems to cut out after 2018 – a bit of searching on their site turns up many more results, although a spot check suggests that at least some of those just have “depression” as a word in the text instead of as a focus. There seems to be a similar situation with bioRxiv. From 2019 onwards, PsyArXiv, OSF Preprints, and Preprints.org seem to be the only real sources.
The above table in graph form:
>> import seaborn as sns
>> sns.set(rc={"figure.figsize": (8,5.5)})
>> sns.histplot(papers_with_good_sources, x="Year", hue="Source",
>> multiple="stack", discrete=True)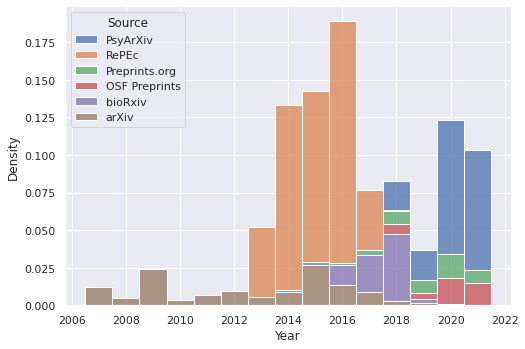
Wrapping Up
The next part is going to be focused on looking at information from the preprints themselves, and potentially diving a bit more into the preprint collections. From the look of it PsyArXiv is going to be the primary source, but bioRxiv, OSF Preprints, and Preprints.org may provide some interesting supplemental material.